Forskjell mellom versjoner av «BioHackerLab/Protokoller»
m (lagt til i kategori Biohacking) |
|||
| (4 mellomliggende revisjoner av en annen bruker er ikke vist) | |||
| Linje 25: | Linje 25: | ||
=DNA quantification= | =DNA quantification= | ||
| + | |||
| + | Reference sample: https://crm.irmm.jrc.ec.europa.eu/p/ERM-AD442k | ||
==Background== | ==Background== | ||
| − | The main methods for determiniing nucleic acid concentrations are UV photometry measuring absorbance at 260 nm (A260) and fluorometry. Note that different sources may describe different conversion factors for conversion from A260 to mass concentration. | + | The main methods for determiniing nucleic acid concentrations are UV photometry measuring absorbance at 260 nm (A260) and fluorometry. Note that different sources may describe different conversion factors for conversion from A260 to mass concentration. For oligomers, a more accurate DNA concentration value may be achieved by estimating the sequence-dependent extinction coefficient by calculation. |
| + | |||
| + | https://www.scripps.edu/california/research/dna-protein-research/forms/biopolymercalc2.html | ||
| + | |||
| + | http://biotools.nubic.northwestern.edu/OligoCalc.html | ||
http://cshprotocols.cshlp.org/content/2007/11/pdb.ip47.full | http://cshprotocols.cshlp.org/content/2007/11/pdb.ip47.full | ||
| Linje 37: | Linje 43: | ||
http://www.ogt.co.uk/resources/literature/483_understanding_and_measuring_variations_in_dna_sample_quality | http://www.ogt.co.uk/resources/literature/483_understanding_and_measuring_variations_in_dna_sample_quality | ||
| + | |||
| + | https://people.rit.edu/rhrsbi/GEPages/LabManualPDF5ed/09%20UV%20Absorption.pdf | ||
| + | |||
| + | https://www.idtdna.com/pages/support/technical-vault/reading-room/quick-reference/quick-reference/2011/06/02/molar-extinction-coefficient | ||
| + | |||
| + | http://www.endmemo.com/bio/OD260.php | ||
=Elektroforese= | =Elektroforese= | ||
| Linje 553: | Linje 565: | ||
BioRad PowerPac Basic Power Supply: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/4006213.pdf | BioRad PowerPac Basic Power Supply: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/4006213.pdf | ||
| + | |||
| + | [[Category:Biohacking]] | ||
Nåværende revisjon fra 6. sep. 2017 kl. 11:29
Innhold
- 1 Resuspendering av DNA-primere
- 2 DNA extraction
- 3 DNA quantification
- 4 Elektroforese
- 5 PCR
- 5.1 Detection of D1S80 Repeat Polymorphism by PCR
- 5.2 Collection of genomic DNA by buccal swabs
- 5.3 PTC tasting ability genotyping
- 5.4 Sex determination by PCR
- 5.5 DNA fingerprinting
- 5.6 Colony PCR
- 5.7 ITS1 + ITS 4 yeast
- 5.8 V9D - LS266 yeast
- 5.9 MEATF + MEATR, animal meats
- 5.10 PORCINE FWD + PORCINE REV, pork
- 6 Lagring av bakterie-stammer i glycerol
- 7 Sekvenseringstjenester
- 8 Brukermanualer
Resuspendering av DNA-primere
Ta røret med tørket DNA ut av pakken. Kontroller at ID og DNA-sekvens oppgitt på røret stemmer med bestillingen.
Plasser røret i en mikrosentrifuge og sentrifuger i ca. 30 sekunder.
Tilsett NF-vann til konsentrasjon etter oppløsning lik 100 pmol/uL (0.1 mM, 100 uM). Se siden av røret eller medfølgende dokumentasjon fra leverandør for total mengde DNA i røret og/eller mengde vann som må tilsettes for endelig konsentrasjon ~100 pmol/uL
Bland ved å knipse på og riste flasken, eller med en vortex-mikser. Sentrifuger til slutt røret igjen.
Oppbevar fryst, fortrinnsvis ved -20C.
Før bruk til PCR vil det være hensiktsmessig å foreta ytterligere 10X fortynning til 10 pmol/uL (10 uM) i et eget rør, som også kan oppbevares fryst og brukes flere ganger. Dette reduserer også behovet for å åpne hovedrøret og reduserer risikoen for kontaminasjon av dette.
Om mulig kontroller konsentrasjonen vha. spektrofotometri. Se http://www.promega.com/a/apps/biomath/index.html?calc=odConvert
DNA extraction
Blount, Driessen & Ellis: GC Preps: Fast and Easy Extraction of Stable Yeast Genomic DNA. Scientific Reports 6, Article number: 26863 (2016). http://www.nature.com/articles/srep26863
Strawberry crude DNA extraction: http://www.stevespanglerscience.com/lab/experiments/strawberry-dna/
DNA quantification
Reference sample: https://crm.irmm.jrc.ec.europa.eu/p/ERM-AD442k
Background
The main methods for determiniing nucleic acid concentrations are UV photometry measuring absorbance at 260 nm (A260) and fluorometry. Note that different sources may describe different conversion factors for conversion from A260 to mass concentration. For oligomers, a more accurate DNA concentration value may be achieved by estimating the sequence-dependent extinction coefficient by calculation.
https://www.scripps.edu/california/research/dna-protein-research/forms/biopolymercalc2.html
http://biotools.nubic.northwestern.edu/OligoCalc.html
http://cshprotocols.cshlp.org/content/2007/11/pdb.ip47.full
http://www.promega.com/a/apps/biomath/
https://people.rit.edu/rhrsbi/GEPages/LabManualPDF5ed/09%20UV%20Absorption.pdf
http://www.endmemo.com/bio/OD260.php
Elektroforese
OBS: Les gjennom hele protokollen før gjennomførelse for å bli kjent med relevant sikkerhetsinformasjon.
Utstyr
OBS: Kontroller utstyret for skader og slitasje før bruk. Ikke fortsett dersom elektroforesekammer, ledninger eller strømforsyning viser tegn til skade.
- Elektroforesekammer. Eks. Carolina Deluxe Gel Chamber.
- Strømforsyning. Eks. BioRad PowerPac Basic
- Automatpipette, 1-10 uL.
- Vekt, 0.1 g eller høyere oppløsning.
- Mikrobølgeovn
- Varmebeskyttende hansker eller annen håndbeskyttelse
- Målesylinder, 50 mL eller større
- Glassflaske til agaroseløsning, 100 mL eller større (må passe i mikrobølgeovn)
- Glassflaske til TAE-bufferløsning, 250 mL eller større
- Transilluminator, eks. DarkReader blue light.
- Stekespade eller lignende for å løfte agarosegel
Avfallsbeholder for pipettespisser og tørt avfall Avfallsbeholder for brukt buffer/flytende avfall
Forbruksvarer:
- Agarose for elektroforese, eks. SeaKem LE agarose. Ca. 0.5 g/gel.
- TAE 10x buffer. Ca. 30 mL.
- Destillert vann: Ca. 300 mL.
- DNA-fargestoff, eks. GelGreen. Ca 5 uL.
- Pipettespisser, 1-10 uL.
- Veieskip.
- Tørkepapir
- DNA ladder
- Loading dye
Støping av agarose-gel
Om nødvendig, lag TAE buffer i brukskonsentrasjon ved å fortynne konsentrert buffer i destillert vann. For å lage 1x TAE buffer fra 10x TAE buffer, bland 1 del TAE buffer og 9 deler destillert vann. TAE buffer er irriterende. Bruk hansker ved håndtering av konsentrert buffer.
Tilsett gradvis ~1 v/v % agarose til 1x TAE buffer (~1g til 100 mL, f.eks 0.5g for 50 mL) i en flaske med volum minst to ganger volumet av løsningen, fortrinnsvis under omrøring med magnetrører - eventuelt rør for hånd med glasstav eller ved å bevege flasken.
OBS: Flasken må ikke være lufttett!
Varm løsningen i mikrobølgeovn på høy styrke til løsningen koker (ca. ett minutt). La løsningen koke i ca. 30 sekunder. Reduser varmeeffekten eller skru av ovnen dersom løsningen koker over.
Fjern flasken fra mikrobølgeovnen. Bland løsningen ved å bevege flasken forsiktig i en roterende bevegelse for hånd, eller med en magnetrører. Utvis forsiktighet for å unngå sprut. Kontroller at agarose-løsningen er klar og uten synlige partikler. Dersom det er partikler og uløst agarose i løsningen, kok løsningen igjen.
OBS: Bruk øyebeskyttelse! Fare for støtkoking og sprut også etter at oppvarmingen har opphørt. Flasken er varm. Bruk varmebeskyttende hansker eller annen håndbeskyttelse ved håndtering av flaske. Bruk fortrinnsvis labfrakk eller langarmede klær og unngå eksponering av bar hud. Fare for forbrenning ved søl eller sprut.
Tilsett DNA-fargestoff i henhold til konsentrasjonsangivelse. F.eks, for 10 000x konsentrert fargestoff, tilsett 1 uL fargestoff per 10 mL agarose løsning (5 uL for 50 mL).
Plasser et gel-støpekar på tvers av lengderetningen i et elektroforesekar og plasser en brønnkam i støpekaret. Hell varm agaroseløsning i kammeret og la stå til gel'en er størknet (ca. 30-60 minutter).
OBS: Elektroforesekaret skal være frakoblet fra strømforsyning når gel'en støpes.
Ta ut brønnkammen og snu støpekaret slik at prøvebrønnene er nærmest den sorte (postive) elektroden.
Hell 1x TAE buffer i elektroforesekaret slik at gel'en er dekket av buffer.
Elektroforese
- Tilsett prøvene i prøvebrønnene.
- Sett på lokket på elektroforesekaret.
- Kontroller at området rundt elektroforesekaret og strømforsyningen er tørt. Tørk bort eventuell væske.
- Koble ledningene til strømforsyningen. Utvis forsiktighet og bruk fortrinnsvis kun en hånd for å redusere risiko for strøm gjennom kroppen.
OBS: Strømforsyningen skal være avslått når ledningene kobles til!
- Skru på strømforsyningen.
- Juster til ønsket spenning og skru på spenningen.
- La elektroforesen foregå uforstyrret. Ikke rør kammeret eller ledningene så lenge spenningen er på. Dersom det oppstår tegn til lekkasje fra kammeret, avslutt elektroforesen umiddelbart uten å komme i kontakt med væsken.
- Skru av spenningen på strømforsyningen.
- Skru av strømforsyningen.
- Koble ledningene fra strømforsyningen. Utvis forsiktighet og bruk fortrinnsvis kun en hånd for å redusere risiko for strøm gjennom kroppen.
- Ta av lokket på elektroforesekaret.
- FLytt agarosegeleen forsiktig til en transilluminator ved hjelp av en stekespade e.l.
- Avhend agarose-gel, bufferløsning og forbruksmateriell på forsvarlig vis etter bruk. Skyll elektroforesekammer og glassutstyr med destillert vann.
Se også: https://www.addgene.org/plasmid-protocols/gel-electrophoresis/
http://www.methodbook.net/dna/agarogel.html
Typisk prøvevolum kan være 6 uL, f.eks 5 uL PCR-produkt + 1 uL 6x loading dye, eller 4 uL vann, 1 uL DNA ladder og 1 uL 6x loading dye.
PCR
Typiske reaksjonsvolum er 20 uL og 50 uL. Gitt en konsentrasjon i primerløsningen lik 10 uM vil i de to tilfellene tilsetting av henholdsvis 1 uL og 2.5 uL av hver primer gi en endelig kosentrasjon av hver primer lik 0.5 uM i reaksjonsblandingen.
http://dongshengbio.com/en/UploadFiles/2012516105050871.pdf
https://www.neb.com/protocols/1/01/01/pcr-protocol-m0530
Typisk vil en ta ut ca. 5 uL av reaksjonsblandingen til elektroforese-analyse etter PCR.
Detection of D1S80 Repeat Polymorphism by PCR
See https://www.dnalc.org/files/pdf/forensicprofchip_d1s80_protocol.pdf
Collection of genomic DNA by buccal swabs
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1566681/pdf/envhper00512-0045.pdf
PTC tasting ability genotyping
https://www.snpedia.com/index.php/Rs713598
https://www.ncbi.nlm.nih.gov/pubmed/12595690?dopt=Abstract
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1181941/
http://www.ncbi.nlm.nih.gov/gene?cmd=retrieve&dopt=default&rn=1&list_uids=5726
http://bioinformatics.dnalc.org/ptc/animation/pdf/ptc.pdf
Sex determination by PCR
See https://en.wikipedia.org/wiki/Amelogenin#Application_in_sex_determination and Fracnes et al.: Clin Chim Acta. 2007 Nov-Dec;386(1-2):53-6. Epub 2007 Jul 31. Amelogenin test: From forensics to quality control in clinical and biochemical genomics.. (http://www.sciencedirect.com/science/article/pii/S0009898107003889)
Primers:
- AME-F: 5'-ctgatggttggcctcaagcctgtg-3'
- AME-R: 5'-taaagagattcattaacttgactg-3'
Expected results:
- If male: 1 band of 977 bp + 1 band of 790 bp
- If female: 1 band of 977 bp
DNA fingerprinting
CODIS
The Combined DNA Index System (CODIS) is a FBI program to support law enforcement by DNA-based identification. CODIS defines standards for DNA fingerprinting and a selection of genetic loci to be analyzed as part of DNA fingerprinting. The 13 CODIS core loci per 2016 are:
- CSF1PO
- FGA
- THO1
- TPOX
- VWA
- D3S1358
- D5S818
- D7S820
- D8S1179
- D13S317
- D16S539
- D18S51
- D21S11
In addition, the Amelogenin (AMEL) locus is used for sex determination.
For information on each locus, see http://www.cstl.nist.gov/strbase/
See also http://www.cstl.nist.gov/strbase/coreSTRs.htm
7 additional loci will be required from January 1 2017.
https://en.wikipedia.org/wiki/Combined_DNA_Index_System
http://www.cstl.nist.gov/biotech/strbase/fbicore.htm
http://www.cybertory.org/resources/CODIS/
Estimated cost for CODIS primer set as described by cybertory.org, with synthesis by Macrogen Inc.: 68.2 EUR / ~75 USD / ~ 650 NOK + shipping.
European set
The European Standard Set (ESS) consists of the following STR loci: FGA, TH01, VWA, D1S1656, D2S441, D3S1358, D8S1179, D10S1248, D12S391, D18S51, D21S11, D22S1045
Additional Loci commonly found in European STR kits include: D2S1338, D16S539, D19S433, SE33, Amelogenin
Colony PCR
1. From an agar plate, select one or several colonies for colony PCR. For each colony selected, pick a small amount of colony material using a sterile pipette tip, and dissolve in 50 uL H2O. Reseed a new agar plate with 5 uL of the resulting solution(s) each in a separate spot, keeping the spots separate and noting the location of each. Alternatively, use 5 uL to inoculate a liquid culture.
Incubate the dissolved colony material at 96 C for 10 min to release DNA.
Use 1 uL of the heat-treated solution as template for PCR.
ITS1 + ITS 4 yeast
Background
See Esteve-Zarzoso et al.: Identification of yeasts by RFLP analysis of the 5.8S rRNA gene and the two ribosomal internal transcribed spacers. (http://www.ncbi.nlm.nih.gov/pubmed/10028278)
Artikkel tilgjengelig via ResearchGate
See also http://sites.biology.duke.edu/fungi/mycolab/primers.htm
PCR setup
According to Esteve-Zarzoso et al.:
Reaction volume: 100 uL
Primer concentrations: 0.5 uM each
Forward (FWD) primer (ITS1): 5' TCCGTAGGTGAACCTGCGG 3'
Reverse (REV) primer (ITS4): 5' TCCTCCGCTTATTGATATGC 3'
(Start and end of expected product sequence = FWD primer + reverse complement of reverse primer: TCCGTAGGTGAACCTGCGG-GCATATCAATAAGCGGAGGA)
Template: Fresh yeast colony material.
Template preparation: 95 C for 15 min.
Initial denaturation: 95C for 5 min
Repeated cycles:
- Denaturation: 94 C for 1 min
- Anneal: 55.5 C for 2 min
- Extension 72C for 2 min
x 35 cycles
Final extension: 72C for 10 min
Total hold time: 190 min
Estimated program duration with OpenPCR: ~4 hr.
Alternative setup and reduced duration PCR program:
Reaction volume: 50 uL
Template: The protocol has been tested with dry yeast, see template preparation below. The protocol may also be attempted with fresh store-bought yeast, liquid yeast culture or yeast agar colony material as the template source.
PCR setup otherwise as original protocol.
Initial denaturation: 94C for 3 min
Repeated cycles:
- Denaturation: 94 C for 30 s
- Anneal: 55.5 C for 30 s
- Extension: 72C for 1 min
x 35 cycles
Step durations and temperatures are according to recommendations for Dongsheng Taq polymerase. Annealing temperature according to original protocol.
Final extension: 72 C for 10 min
Total hold time: 83 min
Run time with OpenPCR: ~ 2h 20 min.
Template preparation:
Dissolve 0.1g dry yeast (Idun tørrgjær) in 10 mL dH20. Mix well. Pipette 50 uL (-> ~0.0005g, 0.5mg yeast powder by dry weight) into a microsentrifuge tube or PCR tube and incubate at 98C for 10 minutes. Use 1-2 uL (-> ~0,00002g = 0,02mg = 20 ug yeast powder by dry weight for 2 uL) of the supernatant as template.
I tørrvekt kan det forventes at DNA utgjør i størrelsesorden 1% av cellemassen. (ref) 20 ug tørrmasse vil da gi anslagsvis ~0.2 ug = 200 ng DNA.
DNA har molekylær vekt lik ca. 650 g/mol per basepar. (ref). S. cerevisae-genomet består av ca. 12,156*106 basepar. Basert på dette får vi da
200 *10-9 g DNA/ 12,156*106 bp) * (650 g/mol * bp) ~ 2,5*10-17 mol genomkopier.
Et mol er lik 6 * 1023. Vi har da
2,5 *10-17 mol * 6 * 1023 / mol ~ 1,5*107 (15 millioner) kopier.
S. Cerevisae har ca. 100-200 kopier av hvert rRNA-gen. Vi får da
15 millioner genomkopier * (~100 genkopier/genom) ~ 1,5 milliarder genkopier.
Extra reduced duration PCR program:
Tested 10.jul.2016. Amplification was successful for 1 of 2 samples. Based on the results, further shortening to 25 cycles may be attempted.
Initial denaturation: 94C for 3 min
Repeated cycles:
- Denaturation: 94 C for 30 s
- Anneal: 55.5 C for 30 s
- Extension: 72C for 1 min
x 30 cycles
No final extension.
Total hold time: 63 min
Run time with OpenPCR: ~112 min. (1 h 52 minutes).
Expected results
Information from literature:
Expected PCR product size for S. cerevisiae (Strains CECT 1942 /ATCC 18824, CECT 1971) as reported by Esteve-Zarzoso et al: 880 bp.
Information from in silico PCR and published genomic sequence data:
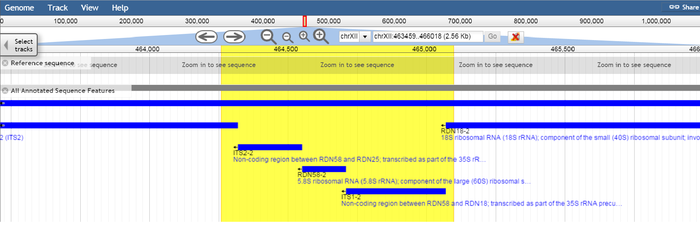
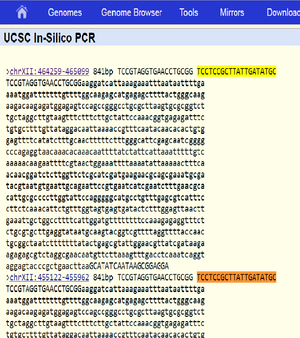
Expected fragment sizes as returned by in silico PCR at https://genome.ucsc.edu/cgi-bin/hgPcr (April 2011 assembly): 2 fragments of 841 bp each (direct link). The reported locations are chrXII:464259-465099 chrXII:455122-455962
The sequences of the two expected products as identified by in silico PCR above are identical. BLAST of the sequence against the R64 Assembly (GenBank GCA_000146045.2) returns two matches, both on chromosome XII: Range 1: 455122 to 455962; and Range 2: 464259 to 465099. Both with 841/841 (100%) identities and zero gaps.
The genome assembly used for the in silico PCR described above is described as "Apr. 2011 (SacCer_Apr2011/sacCer3)". Although the relation or differences between this assembly/description and the R64 assembly have not been determined, the result of the BLAST search shows that the nucleotide positions on chromsome XII as reported in the in silico PCR result correspond to those of the R64 genome assembly and that there are no differences in the sequences of these regions. (direct link to search query)
(The sequence for S288C chromosome XII alone is accesible at: http://www.ncbi.nlm.nih.gov/nuccore/NC_001144.5)
Size & location of expected fragment 1, shown as highlighted area in SGD genome browser.
Size & location of expected fragment 2, shown as highlighted area in SGD genome browser
The target region spanning nucleotide positions 455122 to 455962 covers the 5.8S ribosomal RNA (rRNA) gene RDN58-1 (455414..455571) and the non-coding regions ITS2-1 and ITS1-1 which flank RDN58-1. Likewise, the target region spanning nucleotide positions 464259 to 465099 covers the 5.8S rRNA gene RDN58-2 (464551..464708) and the flanking non-coding regions ITS2-2 and ITS1-2.
SGD entry for RDN58-1: http://www.yeastgenome.org/locus/S000006488/overview
SGD entry for RDN58-2: http://www.yeastgenome.org/locus/S000006489/overview
RDN58-1 and RDN58-2 are contained within the RDN1 locus. Although the sequence included in assembly R64 only indicates two target regions for the ITS1-ITS4 primer combination, the RDN1 locus in actuality represents a 1-2Mbp repeating region containing 100-200 rDNA repeats (ref). Thus the actual number of targets can be expected to be in this range, and some sequence variation among these is likely (ref).
Expected PCR product sequence (841 bp): (NB: Whitespace is included below for easier viewing, adding extra characters)
Esteve-Zarzoso et al. used the restriction enzymes CfoI, HaeIII, and HinfI to cleave PCR product DNA and perform Restriction Fragment Length Polymorphism (RFLP) analysis. The sequence below contains 3 binding sites for CfoI (GCGC) CfoI, 3 binding sites for HaeIII (GGCC) and 3 binding sites for HinfI (GANTC, where N is any nucleotide). The sequence also contains an EcoRI binding site (GAATTC, bolded).
Esteve-Zarzoso reports the restriction fragment sizes for digestion with CfoI, HaeIII and HinfI as 385 + 365 (= 750), 320 + 230 + 180 + 150 (= 880) and 365 + 155 ( = 520), respectively (fragments smaller than 50 bp not included or reported).
Based on the sequence below, the expected fragment sizes from digestion are (http://www.restrictionmapper.org/):
- EcoRI 469 + 372 (= 841)
- CfoI: 363 + 334 + 134 + 10 (= 841)
- HaeIII: 311 + 230 + 172 + 128 (= 841)
- HinfI: 362 + 355 + 116 + 8 ( = 841)
TCCGTAGGTGAACCTGCGGaaggatcattaaagaaatttaataattttga
aaatggatttttttgttttggcaagagcatgagagcttttactgggcaag
aagacaagagatggagagtccagccgggcctgcgcttaagtgcgcggtct
tgctaggcttgtaagtttctttcttgctattccaaacggtgagagatttc
tgtgcttttgttataggacaattaaaaccgtttcaatacaacacactgtg
gagttttcatatctttgcaactttttctttgggcattcgagcaatcgggg
cccagaggtaacaaacacaaacaattttatctattcattaaatttttgtc
aaaaacaagaattttcgtaactggaaattttaaaatattaaaaactttca
acaacggatctcttggttctcgcatcgatgaagaacgcagcgaaatgcga
tacgtaatgtgaattgcagaattccgtgaatcatcgaatctttgaacgca
cattgcgccccttggtattccagggggcatgcctgtttgagcgtcatttc
cttctcaaacattctgtttggtagtgagtgatactctttggagttaactt
gaaattgctggccttttcattggatgttttttttccaaagagaggtttct
ctgcgtgcttgaggtataatgcaagtacggtcgttttaggttttaccaac
tgcggctaatctttttttatactgagcgtattggaacgttatcgataaga
agagagcgtctaggcgaacaatgttcttaaagtttgacctcaaatcaggt
aggagtacccgctgaacttaaGCATATCAATAAGCGGAGGA
V9D - LS266 yeast
Background
"The primers V9D (5'-TTAAGTCCCTGCCCTTTGTA-3') and LS266 (5'-GCATTCCCAAACAACTCGACTC-3') are used [to] amplify an 800-1300 bp fragment that encompasses a portion of the 18S and 28S rRNA genes and the entire intervening ITS1, 5.8S and ITS2 rRNA gene regions. These primers bind to conserved regions, with corresponding positions to Saccharomyces cerevisiae 18S (1609-1627) and 28S (287-266) rRNA genes." (Todd M Pryce. "Universal Detection and Identification of Fungi by PCR and DNA sequencing" in PCR for Clinical Microbiology, SpringerLink 2010.)
PCR setup
As described by Pryce (2010):
- Reaction volume: 50 uL
- Initial denaturation: 95 C for 9 min
- Repeated cycles:
- Denaturation: 95 c for 30s
- Anneal: 62C for 60s
- Extension: 72 C for 2 min
- Final extension: 72C for 5 min
x 33 (PCRS-B variant) or 35 (PCRS-D variant) cycles.
Total hold time: 136,5 min
Expected results
Expected fragment size: 1228 bp. (direct link)
Expected fragment sizes after digestion by EcoRI: 628 + 600.
Expected PCR product sequence (S. cerevisiae):
TTAcGTCCCTGCCCTTTGTAcacaccgcccgtcgctagtaccgattgaat
ggcttagtgaggcctcaggatctgcttagagaagggggcaactccatctc
agagcggagaatttggacaaacttggtcatttagaggaactaaaagtcgt
aacaaggtttccgtaggtgaacctgcggaaggatcattaaagaaatttaa
taattttgaaaatggatttttttgttttggcaagagcatgagagctttta
ctgggcaagaagacaagagatggagagtccagccgggcctgcgcttaagt
gcgcggtcttgctaggcttgtaagtttctttcttgctattccaaacggtg
agagatttctgtgcttttgttataggacaattaaaaccgtttcaatacaa
cacactgtggagttttcatatctttgcaactttttctttgggcattcgag
caatcggggcccagaggtaacaaacacaaacaattttatctattcattaa
atttttgtcaaaaacaagaattttcgtaactggaaattttaaaatattaa
aaactttcaacaacggatctcttggttctcgcatcgatgaagaacgcagc
gaaatgcgatacgtaatgtgaattgcagaattccgtgaatcatcgaatct
ttgaacgcacattgcgccccttggtattccagggggcatgcctgtttgag
cgtcatttccttctcaaacattctgtttggtagtgagtgatactctttgg
agttaacttgaaattgctggccttttcattggatgttttttttccaaaga
gaggtttctctgcgtgcttgaggtataatgcaagtacggtcgttttaggt
tttaccaactgcggctaatctttttttatactgagcgtattggaacgtta
tcgataagaagagagcgtctaggcgaacaatgttcttaaagtttgacctc
aaatcaggtaggagtacccgctgaacttaagcatatcaataagcggagga
aaagaaaccaaccgggattgccttagtaacggcgagtgaagcggcaaaag
ctcaaatttgaaatctggtaccttcggtgcccgagttgtaatttggagag
ggcaactttggggccgttccttgtctatgttccttggaacaggacgtcat
agagggtgagaatcccgtgtggcgaggagtgcggttctttgtaaagtgcc
ttcgaaGAGTCGAGTTGTTTGGGAATGC
MEATF + MEATR, animal meats
Background
Lago et al. describe amplification and sequencing of the cytochrome b (cyt b) gene of several species using a set of degenerate primers. (European Food Research and Technology March 2011, Volume 232, Issue 3, pp 509-515. http://link.springer.com/article/10.1007/s00217-010-1417-1)
PCR setup
Primers:
- MEAT F: CGAGGCCTMTAYTAYGG
- MEAT R: ATTGAKCGTAGGATTGCGTA
M denotes A or C. Y denotes C or T.
PCR program:
- Initial denaturation: 95C for 3 min
- Denaturation: 95 C for 30s
- Annealing: 50C for 30s
- Extension: 72 C for 30s
x 35 cycles.
- Final extension: 72C for 3 min
Expected results
Lago et al. report that DNA amplification with the primers MEAT F/R gave an amplicon of 555 bp in all tested species.
PORCINE FWD + PORCINE REV, pork
Background
See Ilhak and Arslan. Identification of Meat Species by Polymerase Chain Reaction (PCR) Technique. Turk. J. Vet. Anim. Sci. 2007; 31(3): 159-163: http://journals.tubitak.gov.tr/veterinary/issues/vet-07-31-3/vet-31-3-3-0601-30.pdf
The porcine primers used by Ilhak and Arslan were designed based on Lahiff (2001). Mol Cell Probes. 2001 Feb;15(1):27-35. Species-specific PCR for the identification of ovine, porcine and chicken species in meta and bone meal (MBM).
See also GenBank accesion AF039170.1: http://www.ncbi.nlm.nih.gov/nuccore/AF039170
PCR setup
Primers:
- Porcine FWD: GCC TAA ATC TCC CCT CAA TGG TA
- Porcine REV: ATGAAAGAGGCAAATAGATTTTCG
- Reaction volume: 50 uL
- 20 pmol of each primer
PCR program:
- Denaturation: 94 C 45 s
- Annealing: 58C 45 s
- Extension: 72C 90 s.
x 30 cycles.
Electrophoresis
15 uL PCR product, 1.5 % agarose at 100 V for 2 h.
Expected results
Ilhak and Arslan report a PCR product size of 212 bp for porcine meat.
Lagring av bakterie-stammer i glycerol
- 1 Prepare a solution of 60 % v/v glycerol in water. (For 25 mL, mix 10 mL water and 15 mL glycerol)
- 2 Add 400 uL 60 % glycerol solution and 800 uL of the culture to be stored in a cryogenic tube.
- 3 Mix thoroughly!
- 4 Place in 5 C refrigerator for 30 min, then move to -80 C freezer.
Sekvenseringstjenester
GATC (lightrun)
Primer specifications:
- Tm 52-58 C
- 17-19 bp
- G or C at 3' end (max 3 Gs or Cs)
- maximum 4 identical sequential bp.
Preparation instructions:
- Add 5 uL template DNA (80-100 ng/uL plasmid DNA or 20-80 ng/uL purified PCR product) and 5 uL primer, 5 uM (5 pmol/uL) in one 1.5 mL tube.
- Drop off at GATC collection point or ship to: GATC Biotech AG. European Custom Sequencing Centre. Gotrfied-Hagen-Strasse 20. 51105 Köln.
Macrogen
Primer specifications:
- 18-25 bp
- 40-60 % GC
- Tm 55-60
Preparation instructions:
- Add 20 uL DNA (100 ng/uL plasmid or 50 ng/uL purified PCR product) to one tube. Add 20µl primer (10 pmol/uL) to a separate tube (?)
- Ship to: Macrogen Europe, IWO, Kamer IA3-195, Meibergdreef 39,1105 AZ Amsterdam Zuid-oost. Netherlands. Attention: J.S .Park
Preparation guide: https://dna.macrogen.com/eng/support/ces/guide/ces_sample_prep.jsp
Sample submission guide: https://dna.macrogen.com/eng/support/ces/guide/ces_sample_submission.jsp
Brukermanualer
BioRad PowerPac Basic Power Supply: http://www.bio-rad.com/webroot/web/pdf/lsr/literature/4006213.pdf